Section 1
Model Overview
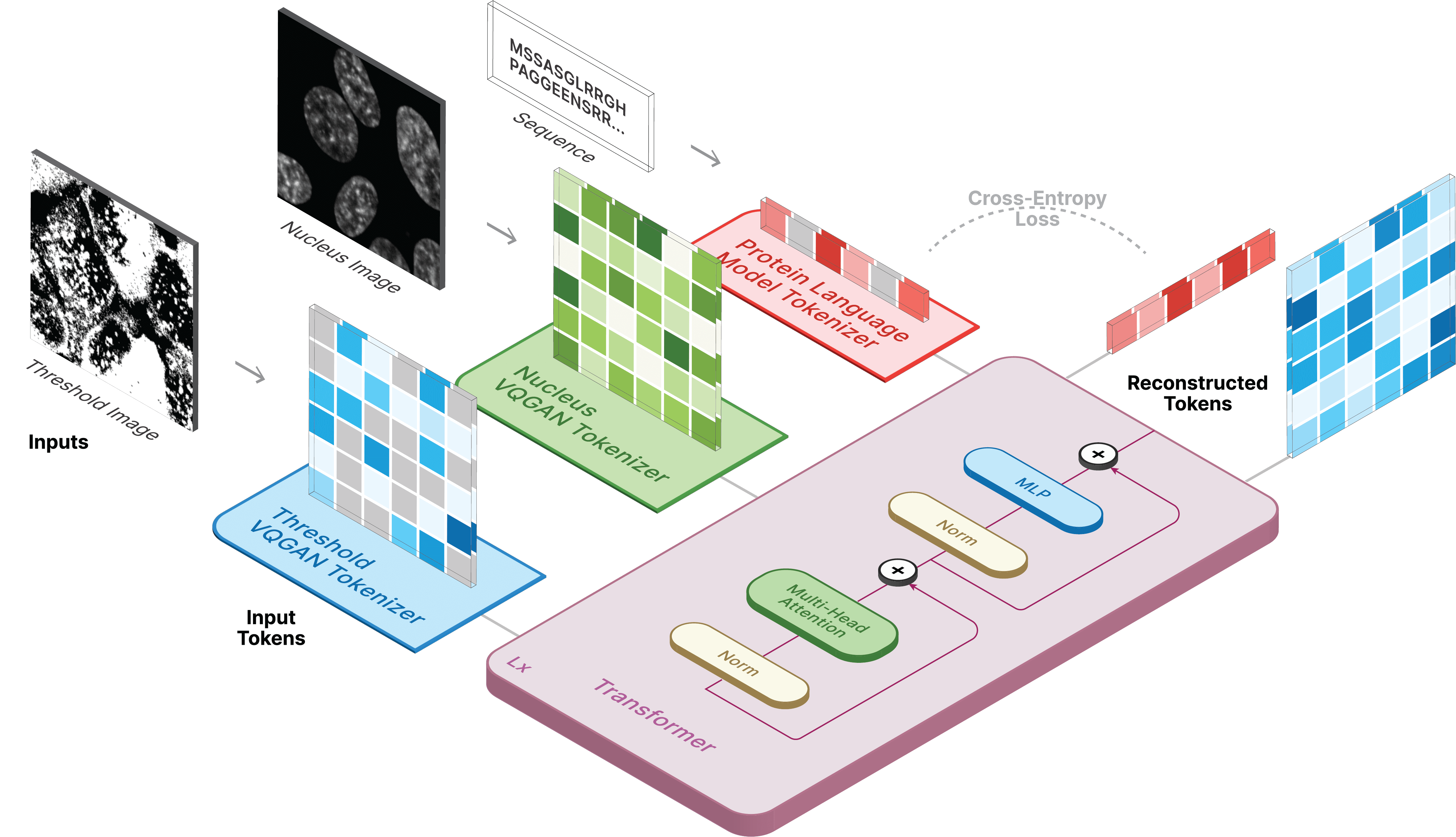
CELL-E 2 is the second iteration of the original CELL-E model which utilizes an amino acid sequence and nucleus image to make predictions of subcellular protein localization with respect to the nucleus.
We use a novel bidirectional transformer that can generate images depicting protein subcellular localization from the amino acid sequences (and vice versa).
CELL-E 2 can not only capture the spatial complexity of protein localization and produce probability estimates of localization atop a nucleus image, but it is also able to generate sequences from images, enabling de novo protein design.
We trained on the Human Protein Atlas (HPA) and the OpenCell datasets. CELL-E 2 utilizes pretrained amino acid embeddings from ESM-2. Localization is predicted as a binary image atop the provided nucleus. The logit values are weighted against these binary images to produce a heatmap of expected localization.
Section 2
Localization Prediction
Text-to-Image
CELL-E 2 can generate localization images by masking the image input section.
Section 3
Sequence Prediction
Image-to-Text
Similarly, amino acids positions can be masked (replaced or inserted) to make predictions based on the localization pattern.
Section 4
De novo Protein Design
We created an entirely new approach to protein design which leverages spatial information from images.
Using CELL-E 2, we predicted 255 likely novel nuclear localizing signals with distinct sequence homology from documented sequences.
Section 5
Comparison
In comparison to CELL-E, CELL-E 2 makes image predictions 65x faster with higher accuracy.